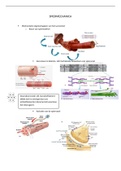
Translation and proteins I (KCSPK 14)
Translations and proteins
1. Translation is the biological polymerization of
amino acids into polypeptide chains
2. Process requires:
- amino acids
- messenger RNA (mRNA)
- ribosomes
- means of directing amino acids into the correct
position
Translation
Ribosomes
1. Converts the message into Macromolecules – “translation machines”
a polypeptide
2. “Language of nucleic acids” 1. Abundant in cell: 10 000 copies in bacterial cells
“language of amino acids” (many more in eukaryotic cells)
2. 20 -30 nm diameter
3. Large & small subunits (monosome = large +
small subunits assembled)
4. rRNA (catalytic function) and proteins (fine-tuning)
– translational apparatus
5. Redundancy in rRNA genes – multiple gene copies
6. Polycistronic transcription of rRNA
- Prokaryotes and eukaryotes
“S: = Svedberg coefficient (unit of measured rate of
ribosomes in a sucrose gradient – associated with
,density, mass and shape of the macromolecule) Note
that “S” is not additive
2. Eukaryotes (mammals):
Polysomes – multiple ribosomes translating the same
- 80S (60S + 40S)
mRNA molecules
- Large subunit: 28S +
5.8S + 5S + 50
Growing polypeptide proteins
chain subunit
Large 3. Small subunit: 18S + 35
Small subunit proteins
mRNA
Ribosomes
1. Prokaryotes (E. coli):
- 70S (50S + 30S)
- Large subunit: 23S +
5S rRNAs + 33
proteins
- Small subunit: 16S +
21
proteins
, Components of pro-and eukaryotic ribosomes tRNA
4. Small (75-90 nucleotides), stable
5. Practically identical in prokaryotes and eukaryotes
6. Transcribed as larger precursor; spliced into 4S
tRNA molecules
7. Holley et al. 1965 established complete nucleotide
sequence of tRNAala
8. Contains unique bases
- Post-transcriptional modifications
- E.g. inosinic acid; ribothymidylic acid and
pseudouridylic acid
Unique bases in
tRNAs formed by
enzymatic
posttranscriptional
modification
Typical cloverleaf structure of
tRNAs
tRNA structure
(2D)
Translations and proteins
1. Translation is the biological polymerization of
amino acids into polypeptide chains
2. Process requires:
- amino acids
- messenger RNA (mRNA)
- ribosomes
- means of directing amino acids into the correct
position
Translation
Ribosomes
1. Converts the message into Macromolecules – “translation machines”
a polypeptide
2. “Language of nucleic acids” 1. Abundant in cell: 10 000 copies in bacterial cells
“language of amino acids” (many more in eukaryotic cells)
2. 20 -30 nm diameter
3. Large & small subunits (monosome = large +
small subunits assembled)
4. rRNA (catalytic function) and proteins (fine-tuning)
– translational apparatus
5. Redundancy in rRNA genes – multiple gene copies
6. Polycistronic transcription of rRNA
- Prokaryotes and eukaryotes
“S: = Svedberg coefficient (unit of measured rate of
ribosomes in a sucrose gradient – associated with
,density, mass and shape of the macromolecule) Note
that “S” is not additive
2. Eukaryotes (mammals):
Polysomes – multiple ribosomes translating the same
- 80S (60S + 40S)
mRNA molecules
- Large subunit: 28S +
5.8S + 5S + 50
Growing polypeptide proteins
chain subunit
Large 3. Small subunit: 18S + 35
Small subunit proteins
mRNA
Ribosomes
1. Prokaryotes (E. coli):
- 70S (50S + 30S)
- Large subunit: 23S +
5S rRNAs + 33
proteins
- Small subunit: 16S +
21
proteins
, Components of pro-and eukaryotic ribosomes tRNA
4. Small (75-90 nucleotides), stable
5. Practically identical in prokaryotes and eukaryotes
6. Transcribed as larger precursor; spliced into 4S
tRNA molecules
7. Holley et al. 1965 established complete nucleotide
sequence of tRNAala
8. Contains unique bases
- Post-transcriptional modifications
- E.g. inosinic acid; ribothymidylic acid and
pseudouridylic acid
Unique bases in
tRNAs formed by
enzymatic
posttranscriptional
modification
Typical cloverleaf structure of
tRNAs
tRNA structure
(2D)