Cells and Immunity
Week 10
Mike Fry
Proteasome and Autophagy
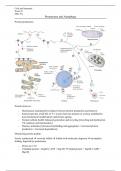
Protein production;
Protein turnover;
- Homeostasis maintained by balance between protein production and turnover
- Each protein has a half life of T ½ [varies between minutes to weeks]; modified by
post translational modifications and protein ageing.
- Normal cellular health; balanced generation and recycling [recycling and dysfunction
VS synthesis and functionality]
- Disease; imbalance [increased misfolding and aggregation // increased protein
production // increased degradation]
Monitoring protein quality;
Newly synthesized correctly folded folded with molecular chaperon incomplete
folding, digested by proteasome.
- Errors are 1/10
- Unfolded protein + Hsp60 [+ATP + Hsp10] folded protein + Hsp60 [+ADP +
Hsp10]
, Cells and Immunity
Week 10
Mike Fry
Degradation of proteins;
- 80-90% in 26S proteasome
- 10-20% in autolysosome with cathepsins
- Proteasome;
o Found in cytosol and nucleus [1%
of cell protein]
o Production regulated by signalling
o Two forms;
1. 20S [alpha and beta subunits],
700kDa, broad substrate
specificity, threonine protease
2. 26S [20S complex with 19S on
either end], 2000kDa
- P53 controlled by regulated destruction;
Post translational modifications;
- Ubiquitylation [attach ubiquitin] / phosphorylation / methylation / acetylation
- Ubiquitylation of substrates;
o Ub is 76 amino acids, mediated by E1 [ubiquitin activating enzyme] / E2
[ubiquitin conjugating enzyme] and E3 [ubiquitin ligase]
o Monoubiquitylation/ multiubiquitylation [one or many Ub attached
separately]; Ub protein interactions/localization/ modulation of protein activity
o Polyubiquitylation [multiple chains of Ub attached] can either act as a target
for 26S proteasome or NF-kB activation/DNA repair/target lysosome
Week 10
Mike Fry
Proteasome and Autophagy
Protein production;
Protein turnover;
- Homeostasis maintained by balance between protein production and turnover
- Each protein has a half life of T ½ [varies between minutes to weeks]; modified by
post translational modifications and protein ageing.
- Normal cellular health; balanced generation and recycling [recycling and dysfunction
VS synthesis and functionality]
- Disease; imbalance [increased misfolding and aggregation // increased protein
production // increased degradation]
Monitoring protein quality;
Newly synthesized correctly folded folded with molecular chaperon incomplete
folding, digested by proteasome.
- Errors are 1/10
- Unfolded protein + Hsp60 [+ATP + Hsp10] folded protein + Hsp60 [+ADP +
Hsp10]
, Cells and Immunity
Week 10
Mike Fry
Degradation of proteins;
- 80-90% in 26S proteasome
- 10-20% in autolysosome with cathepsins
- Proteasome;
o Found in cytosol and nucleus [1%
of cell protein]
o Production regulated by signalling
o Two forms;
1. 20S [alpha and beta subunits],
700kDa, broad substrate
specificity, threonine protease
2. 26S [20S complex with 19S on
either end], 2000kDa
- P53 controlled by regulated destruction;
Post translational modifications;
- Ubiquitylation [attach ubiquitin] / phosphorylation / methylation / acetylation
- Ubiquitylation of substrates;
o Ub is 76 amino acids, mediated by E1 [ubiquitin activating enzyme] / E2
[ubiquitin conjugating enzyme] and E3 [ubiquitin ligase]
o Monoubiquitylation/ multiubiquitylation [one or many Ub attached
separately]; Ub protein interactions/localization/ modulation of protein activity
o Polyubiquitylation [multiple chains of Ub attached] can either act as a target
for 26S proteasome or NF-kB activation/DNA repair/target lysosome