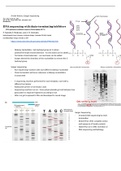
A brief history: Sanger Sequencing
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC431765/
- Dideoxy Nucleotides - lack hydroxyl group at 3 carbon
positionTerminate strand extension –no new bases can be added
- Terminate strand extension – no new bases can be added
- Sanger altered the chemistry of the nucleotides to remove the 3’
hydroxyl group
Sanger sequencing:
- Run sequencing reactions with each different dideoxy-nucleotide
- Chain termination will occur whenever a dideoxy-nucleotide is
incorporated
- 4 sequencing reactions performed for each template, each with a
different terminator
- Radioactive primer or terminator used
- Sequencing reactions run on <1mm polyacrylamide gel cast between two
glass plates to separate fragments according to size
- After run gel is exposed to film and developed to reveal image
Sanger Sequencing
- Invented DNA sequencing by chain
termination
- (Nobel Prize 1958: complete amino
acid sequence of insulin protein)
- Nobel Prize 2(!!) 1980: Invention of
DNA sequencing methodology
, Sanger sequencing
- Radio-labelled chain terminating dideoxy-nucleotides
- Stop primer extension when a specific nucleotide is added
- Read DNA sequences by running samples on a gel
Combinations of different technologies:
- Fluorescent chemistry
- Laser technology
- Imaging
- Data handling and storage
- Computing power
Fluorescent chain terminators:
- Limited to sequencing one sample at a time
- Still gel based
- Still incredibly labour intensive
- Eepensive: ABI 3730, 0.001Gb/run, $1,000,000 per Gb
- 1Gb = 1 billion nucleotides = 1,000,000,000
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC431765/
- Dideoxy Nucleotides - lack hydroxyl group at 3 carbon
positionTerminate strand extension –no new bases can be added
- Terminate strand extension – no new bases can be added
- Sanger altered the chemistry of the nucleotides to remove the 3’
hydroxyl group
Sanger sequencing:
- Run sequencing reactions with each different dideoxy-nucleotide
- Chain termination will occur whenever a dideoxy-nucleotide is
incorporated
- 4 sequencing reactions performed for each template, each with a
different terminator
- Radioactive primer or terminator used
- Sequencing reactions run on <1mm polyacrylamide gel cast between two
glass plates to separate fragments according to size
- After run gel is exposed to film and developed to reveal image
Sanger Sequencing
- Invented DNA sequencing by chain
termination
- (Nobel Prize 1958: complete amino
acid sequence of insulin protein)
- Nobel Prize 2(!!) 1980: Invention of
DNA sequencing methodology
, Sanger sequencing
- Radio-labelled chain terminating dideoxy-nucleotides
- Stop primer extension when a specific nucleotide is added
- Read DNA sequences by running samples on a gel
Combinations of different technologies:
- Fluorescent chemistry
- Laser technology
- Imaging
- Data handling and storage
- Computing power
Fluorescent chain terminators:
- Limited to sequencing one sample at a time
- Still gel based
- Still incredibly labour intensive
- Eepensive: ABI 3730, 0.001Gb/run, $1,000,000 per Gb
- 1Gb = 1 billion nucleotides = 1,000,000,000