1.1 PRINCIPLES OF DNA CLONING
1.1.1 CELL BASED DNA CLONING
4 steps of cell based DNA cloning :
1) In vitro construction of a recombinant DNA molecule
2) Transformation in a host
3) Selective propagation of clones
4) Isolation of recombinant DNA clones
STEP 1 : IN VITRO CONSTRUCTION OF A RECOMBINANT DNA MOLECULE
Recombinant DNA molecule = combining different DNA molecules
→ requires cutting and pasting of DNA
● Cutting = restriction endonucleases
● Pasting = DNA ligase
Requires a replicon
● A piece of DNA that makes independent DNA replication possible
● Region of an organism’s genome that independently replicates from a single origin of replication (ORI)
● Replicon is specific for a host
Plasmid
● Mostly we clone in bacteria → replicon = plasmid
● Plasmid is a small circular DNA molecule found in bacteria
● They are physically separated from chromosomal DNA and replicate independently
Vector
● Usually a construct called “vector” is used, containing many features used in the cloning process
● Vector = plasmid that has been engineered → special features have been added
● It’s used as a vehicle to carry a particular DNA segment into a host cell
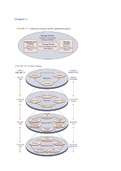
Schema
1) Bacterial cells with vector = plasmid
● Purify plasmid
● Cut it with restriction enzyme
2) Human cells
● Purify DNA
● Cut it with restriction enzyme
3) Ligate bacterial DNA with human DNA = recombinant DNA
,STEP 2 : TRANSFORMATION
Transformation = recombinant DNA molecule is introduced in a host cell
● Usually introduction in bacterial cells or yeast
○ Easy to grow
○ Fast reproduction
● For expression studies, cloning is often done in eukaryotic cells
○ Mammalian cells, insect cells → see later in this chapter
● So take construct → put it in host cells = transformation
STEP 3 : SELECTIVE PROPAGATION OF CLONES
● The transformed cells are plated on agar → each individual cell forms a colony
● 1 colony can be grown in liquid medium to obtain more cells
● Each colony consists of clones
○ All cells are identical because they have the same ancestor cell
STEP 4 : ISOLATION OF RECOMBINANT DNA CLONES
Cell lysis and purification
● 1 colony is picked up from the agar and will be grown in liquid medium → grow it overnight → liquid
medium with a lot of bacteria → lyse them → purify recombinant DNA
→ now look back for more details
,1.1.2 RESTRICTION ENDONUCLEASES
→ First step : making recombinant DNA molecule
Restriction endonucleases (RE) = enzymes used for cutting to make recombinant DNA
● Nomenclature: 1 letter genus, 2 letters species, followed by number
○ E.g. HaeIII: Hemophilus aegypticus
○ The RE is named after a bacteria because we get these enzymes from bacteria
○
Function of RE in bacteria = defense mechanism against bacteriophages
● Bacteria can get infected by viruses = bacteriophages
● DNA from bacteriophage enters the bacterium and is cleaved
● Type II RE will cut a specific recognition sequence
● RE recognises a specific DNA sequence BUT we don’t want the bacterial DNA to be cut
○ There’s a matching sequence specific DNA methylase
○ It methylases the recognition sequence in the bacterial DNA → bacterial DNA cannot be cut
Recognition sequence
● Usually 4-8 bp
● Usually palindrome
Cleavage
● On the symmetry axis: blunt ends
● Not on the symmetry axis : overhangs = sticky ends = cohesive termini
● Sticky ends can base pair and form unstable double helices by DNA ligase
○ 5’ overhang
○ 3’ overhang
, Examples of restriction enzymes that are used in the lab
MBO1 and BAMH1 create the same overhangs
● 5 prime overhang = GATC
● These overhangs are compatible = They can be ligated
Examples of RE and the results they give on total human DNA
➔ Take the whole human genome and cut it with an RE = you get fragments of a certain length (bp)
➔ If you have a short recognition sequence you get short fragments
1) HaeIII gives longer fragments than AluI, even though the recognition sequence is equal in length (4 bp)
● The human genome has less GC than AT
● There will be less sites for HAEIII than for AluI
● Therefore the fragments of AluI will be shorter than HAEIII fragments
2) TaqI gives even longer fragments
● It has the same GC content than AluI but gives longer fragments
● Has to do with CpG → important site for DNA methylation
● CpG are only ⅕ of the frequence of a dinucleotide = rare in the human genome
Isoschizomers: different restriction enzymes that have the same recognition sequence
Some RE have compatible cohesive termini → E.g. BamHI: GGATCC en MboI: GATC